Draw Phylogenetic Tree
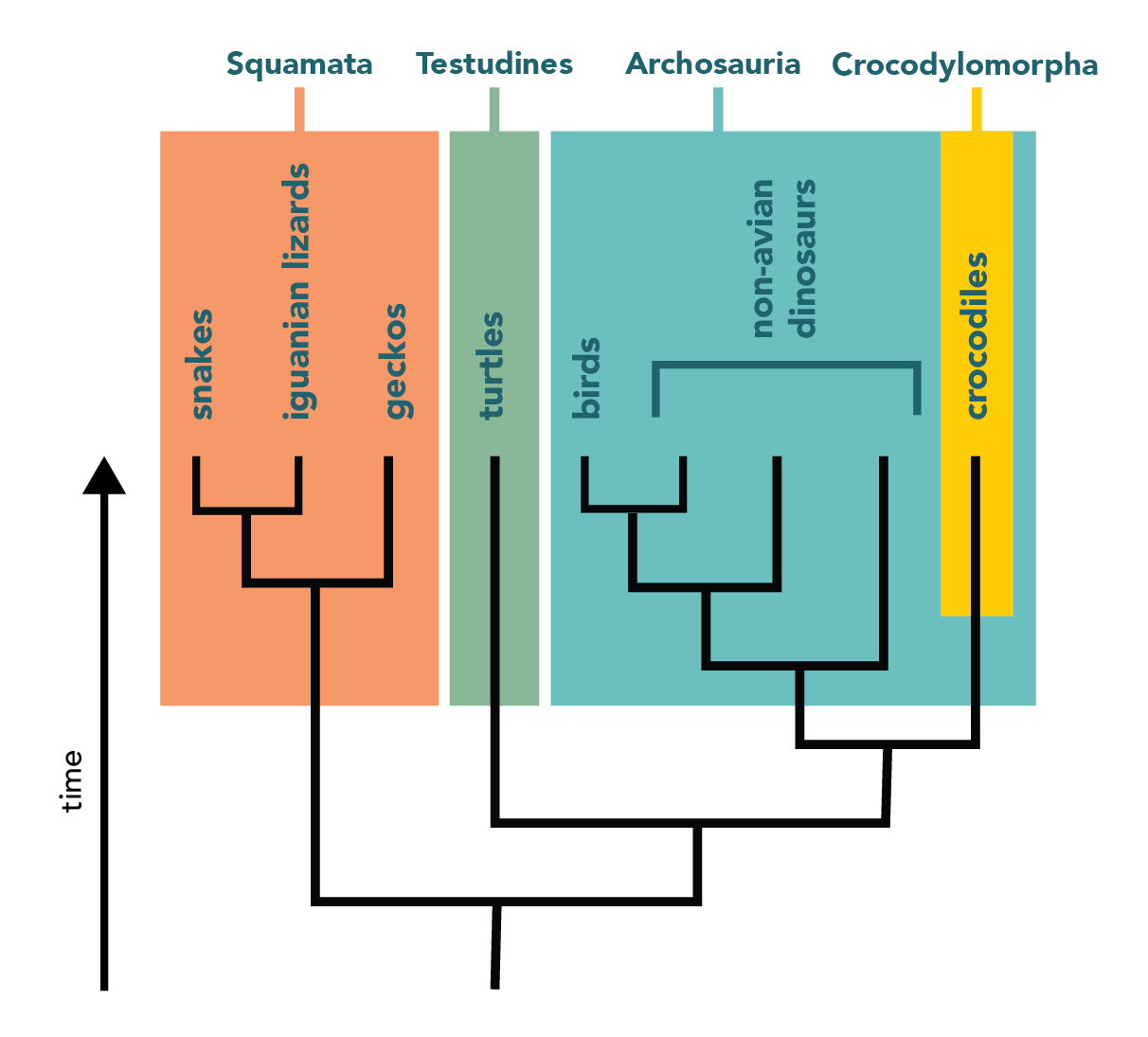
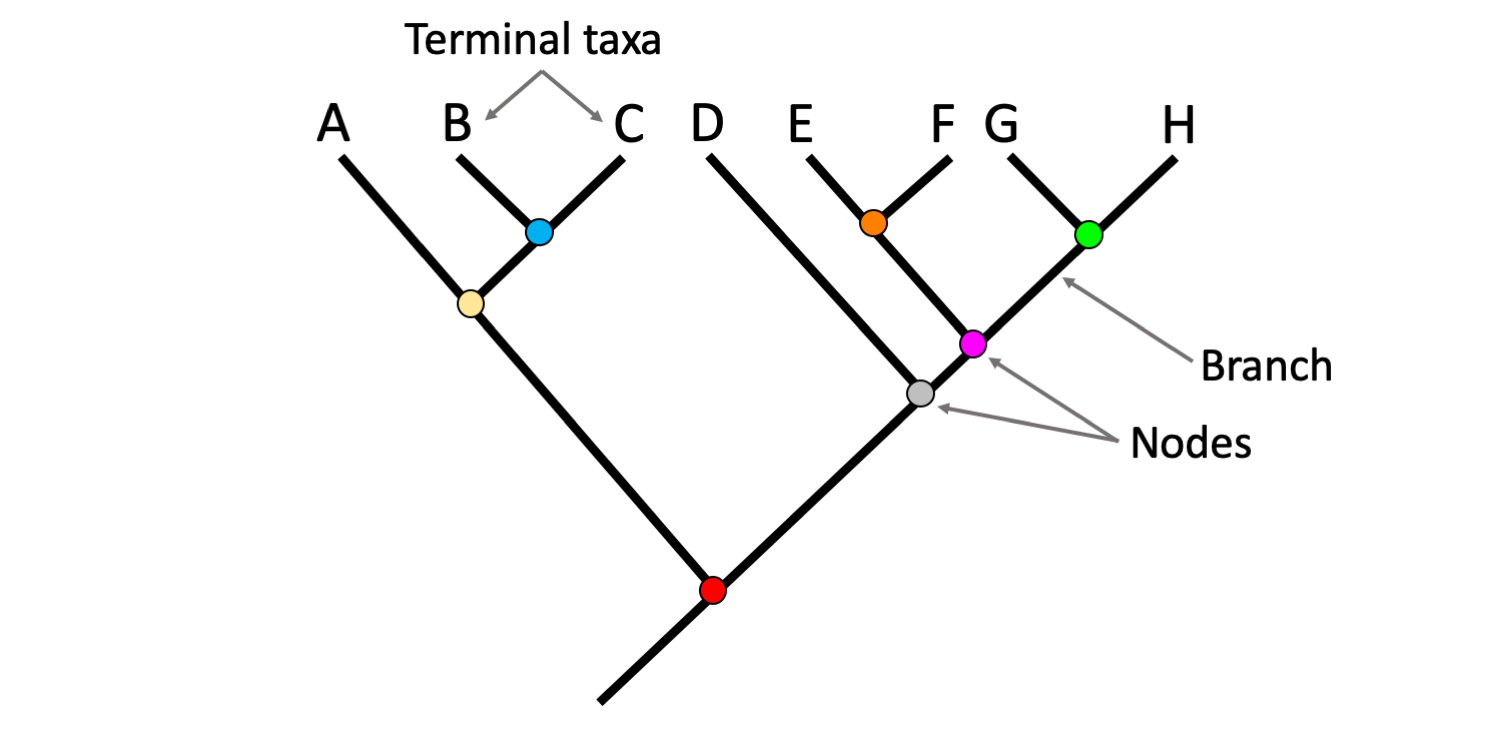
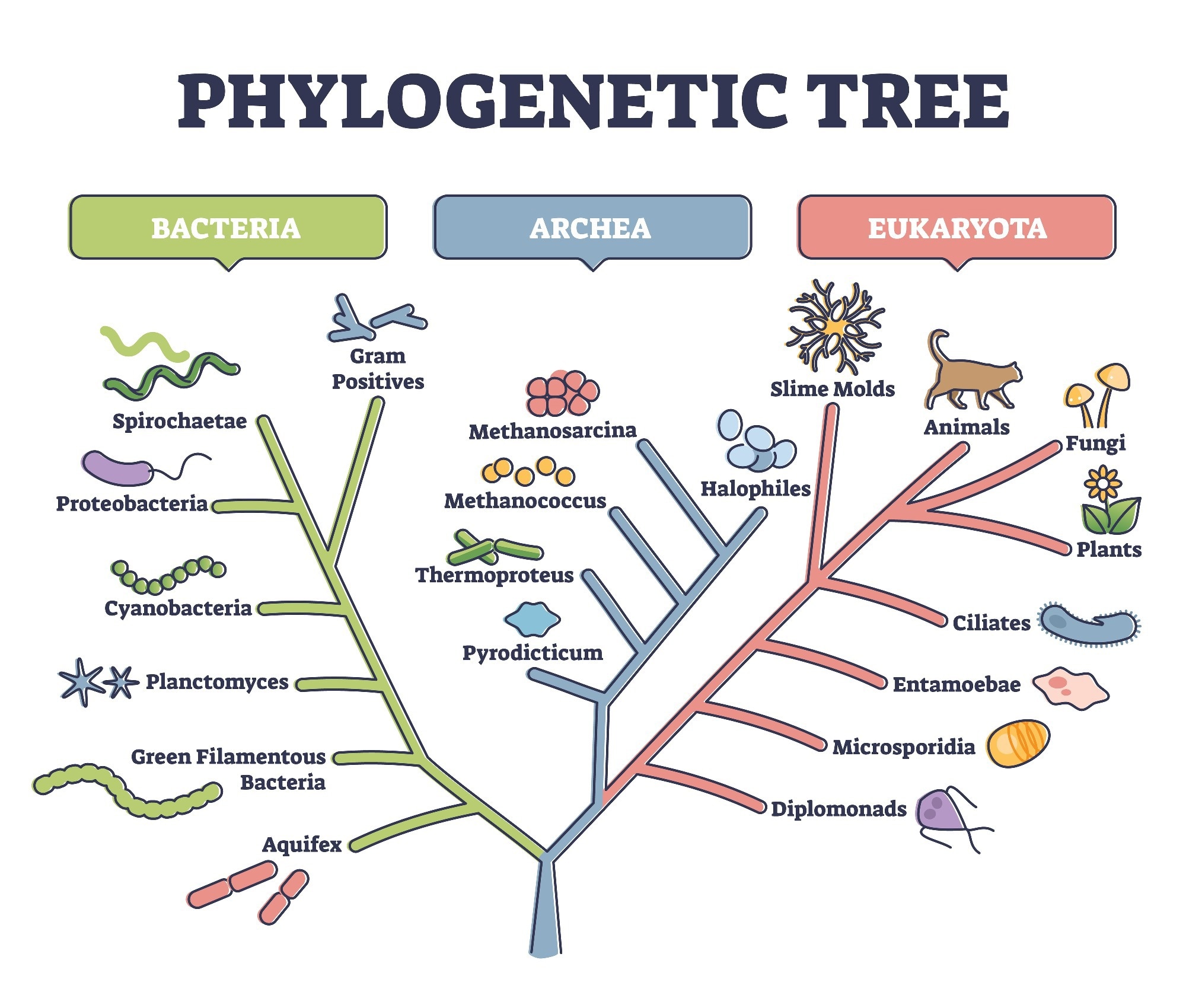
Draw Phylogenetic Tree - You can run codeml and slr easly using ete, as well as visualizing results. Hybridization and genome polyploidy are the main driving forces of genome evolution and the basis of heterosis and polyploidy, which greatly enriches the species diversity in nature. From a list of taxonomic names, identifiers or protein accessions, phylot will generate a pruned tree in the selected output format. In most cases, researchers draw phylogenetic trees in such a way as to record only those events that are relevant to a set of living. It is often referred to as the “number one forest oxygen bar in southern jiangsu.” for urban dwellers seeking respite from the hustle and bustle, baoyinghu national wetland park provides a breath of fresh. In building a tree, we organize species into nested groups based on shared derived traits (traits different from those of the group's ancestor). Manage and visualize your trees directly in the browser, and annotate them with various datasets. Web interactive tree of life is an online tool for the display, annotation and management of phylogenetic and other trees. Rootroot and rearrange the trees. Web treegraph 2 is a is graphical editor for phylogenetic trees, which allows to apply various of graphical formats and edit operations and supports several (visible or invisible) annotations attached to nodes or branches. [1] [2] in other words, it is a branching diagram or a tree showing the evolutionary relationships among various biological species or other entities based upon. Annotate trees with taxonomic data. For more complete documentation, see the phylogenetics chapter of the biopython tutorial and the bio.phylo api pages generated from the source code. Web the park covers an expanse of. Automated reconstruction of gene and species trees. Dynamic connectors to easily draw evolutionary connections. For more complete documentation, see the phylogenetics chapter of the biopython tutorial and the bio.phylo api pages generated from the source code. Trees can represent relationships ranging from the entire history of life on earth, down to individuals in a population. Run phylogenetic workflows with ease. Start practicing—and saving your progress—now: Use the ncbi taxonomy database to perform queries efficiently or to annotate your. Trees can represent relationships ranging from the entire history of life on earth, down to individuals in a population. Web this is an online tool for phylogenetic tree view (newick format) that allows multiple sequence alignments to be shown together with the. For a free version without save/export function use. Based on the phylogenetic tree above, what species is most related to the blue whale? Hybridization and genome polyploidy are the main driving forces of genome evolution and the basis of heterosis and polyploidy, which greatly enriches the species diversity in nature. Run phylogenetic workflows with ease. Manage and visualize your trees. Web example question #1 : Nodes are a visual tool used to separate two groups. Web in this article, we'll take a look at phylogenetic trees, diagrams that represent evolutionary relationships among organisms. Use the ncbi taxonomy database to perform queries efficiently or to annotate your. This interactive module shows how dna sequences can be used to infer evolutionary relationships. From a list of taxonomic names, identifiers or protein accessions, phylot will generate a pruned tree in the selected output format. Rootroot and rearrange the trees. It uses the tree drawing engine implemented in the ete toolkit, and offers transparent integration with the ncbi taxonomy database. Based on the phylogenetic tree above, what species is most related to the blue. Web a phylogenetic tree is a visual representation of the relationship between different organisms, showing the path through evolutionary time from a common ancestor to different descendants. For a free version without save/export function use. What does a node represent on a phylogenetic tree? Web example question #1 : Web phylot generates phylogenetic trees based on the ncbi taxonomy or. Swipe left/right to switch between tree types. Web scientists use a tool called a phylogenetic tree, a type of diagram, to show the evolutionary pathways and connections among organisms. By practicing parsimony, we aim for the simplest explanation. Scientists can estimate these relationships by studying the organisms’ dna. Web treegraph 2 is a is graphical editor for phylogenetic trees, which. Hybridization and genome polyploidy are the main driving forces of genome evolution and the basis of heterosis and polyploidy, which greatly enriches the species diversity in nature. This interactive module shows how dna sequences can be used to infer evolutionary relationships among organisms and represent them as phylogenetic trees. From a list of taxonomic names, identifiers or protein accessions, phylot. Automated reconstruction of gene and species trees. The most recent common ancestor between the two branches. Web a phylogenetic tree, phylogeny or evolutionary tree is a graphical representation which shows the evolutionary history between a set of species or taxa during a specific time. Our main products include home glass decoration, garden decoration and festival glass ornament for christmas, halloween.. Cotton, wheat, rape and other major grain, cotton and oil crops are typical allopolyploid species. In most cases, researchers draw phylogenetic trees in such a way as to record only those events that are relevant to a set of living. Dynamic connectors to easily draw evolutionary connections. Annotate trees with taxonomic data. Web courses on khan academy are always 100% free. This interactive module shows how dna sequences can be used to infer evolutionary relationships among organisms and represent them as phylogenetic trees. Web example question #1 : Draw and interpret phylogenetic trees. Nodes are a visual tool used to separate two groups. [1] [2] in other words, it is a branching diagram or a tree showing the evolutionary relationships among various biological species or other entities based upon. Data can be imported from many tree formats, tables and bayestraits output. Swipe left/right to switch between tree types. This module provides classes, functions and i/o support for working with phylogenetic trees. It is often referred to as the “number one forest oxygen bar in southern jiangsu.” for urban dwellers seeking respite from the hustle and bustle, baoyinghu national wetland park provides a breath of fresh. Web scientists use a tool called a phylogenetic tree, a type of diagram, to show the evolutionary pathways and connections among organisms. Web freehand drawing and highlights to sketch as you detail and analyze organisms.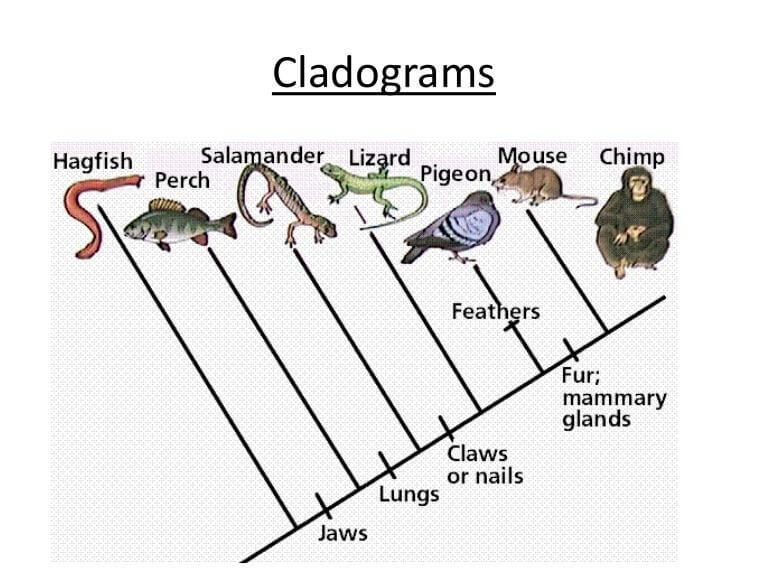
Trees Explained with Examples EdrawMax
Using the tree for classification Understanding Evolution
tree generator from table Ardith Galarza

drawing a tree YouTube

A simplified tree of the kingdom Animalia showing only the

Tree Template
What is Molecular

trees in R using ggtree The Molecular Ecologist
[Solved] How do you construct a tree for the data table
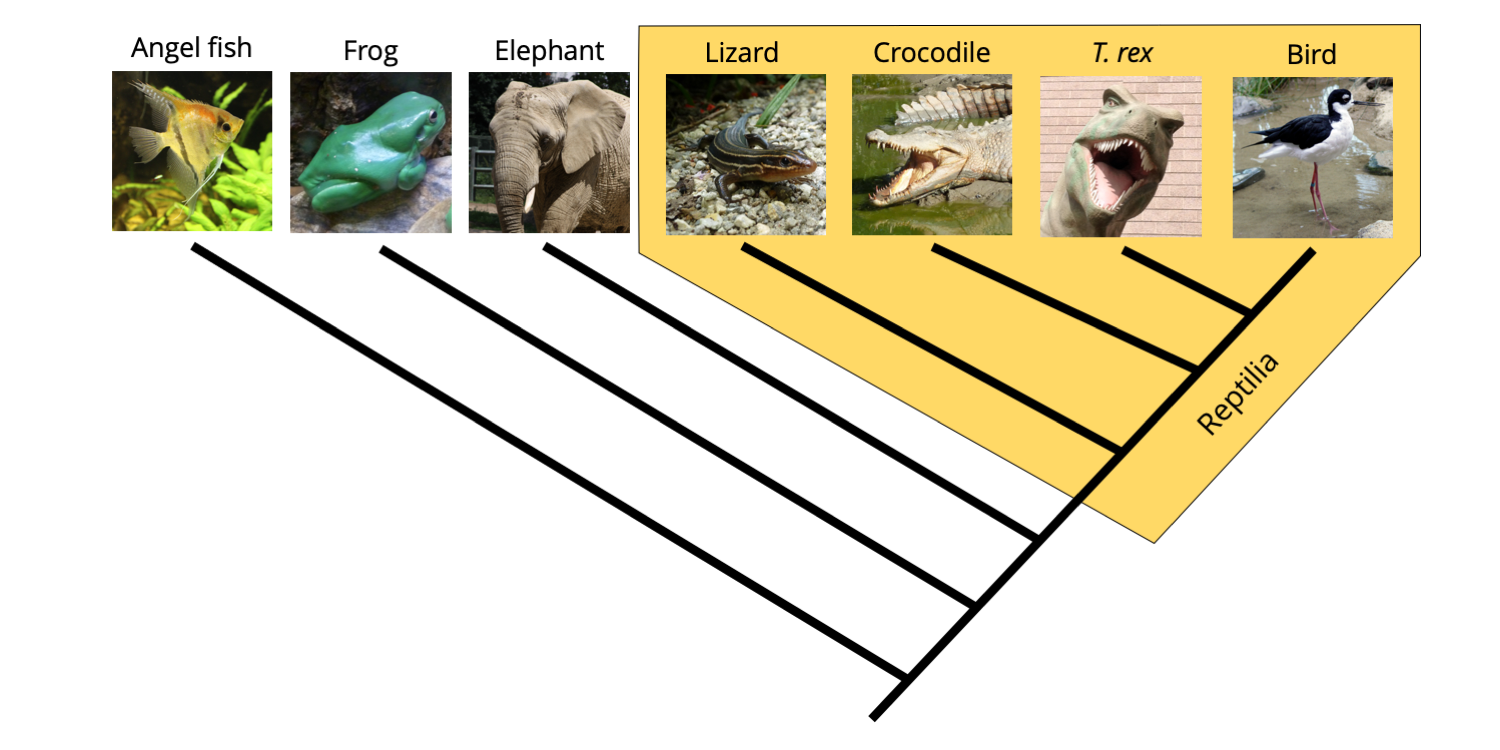
2.4 Trees and Classification Digital Atlas of Ancient Life
Trees Can Represent Relationships Ranging From The Entire History Of Life On Earth, Down To Individuals In A Population.
Hybridization And Genome Polyploidy Are The Main Driving Forces Of Genome Evolution And The Basis Of Heterosis And Polyploidy, Which Greatly Enriches The Species Diversity In Nature.
Vertical Zoom Gesture To Enlarge Plain Tree.
Web A Phylogenetic Tree, Phylogeny Or Evolutionary Tree Is A Graphical Representation Which Shows The Evolutionary History Between A Set Of Species Or Taxa During A Specific Time.
Related Post: